
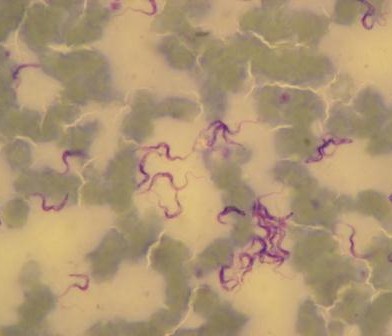
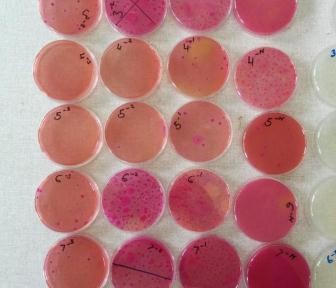
From the ongoing case control study at both Korogocho and Viwandani study sites, a total of 327 samples have been analyzed {water 146; food 134 and surface swabs 47} at University of Nairobi, Department of Public Health, Pharmacology and Toxicology. Of the samples analyzed 67% (219) had bacteria growth {97 water on Aqua 3M petri film plates, 96 food and 26 swab samples on 3M enterobacteriaceae petri film plates}. The study has so far isolated, identified and stocked Escherichia coli (109), Salmonella spp(8) and Shigella spp(6) from 108 samples.
Two hundred sixty seven (267) isolates have not been identified yet and will require the use of the ‘API system’. Antimicrobial sensitivity tests for the identified isolates, at both phenotypic and genotypic levels are yet to be undertaken, due to delayed laboratory supplies. The isolates both identified and those waiting more tests are currently being stored in cryogenic vials at ‐200C in glycerol/skimmed milk. There has been recruitment of 4 Technologists with the retainment of three (3) after one moved for permanent employment elsewhere. The Technologists underwent hands‐on training at University of Nairobi and KEMRI laboratories and currently handling laboratory samples and carrying out bacteria isolation and identification.
The project has three MSc students: ‐
- Aondo Ezra Ochami : Prevalence and risk factors associated with pathogenic Escherichia coli infection of children in selected slum areas of Nairobi, Kenya
- Mercy Cianjoka Gichuyia: Prevalence and antimicrobial resistance of Salmonella species isolated from food, livestock and the environment in Korogocho and Viwandani slums, Nairobi, Kenya.
- Macharia James : A study of antimicrobial resistance of Escherichia coli isolated from livestock and the environment in Korogocho and Viwandani ,Nairobi, Kenya.
SUMMARY REPORT FROM KEMRI LAB
The stool samples are collected from children in Viwandani and Korogocho and brought to CMR/KEMRI Laboratory for Rotavirus, Parasitic and bacterial analysis. In summary, the processing of samples in the lab involves 1) parasitological 2) bacteriological and Rotavirus analysis. So far, 198 samples have been collected. For bacteriological tests, 88.9% are positive for E. coli while on parasitology tests, 18% are positive for Giardia lamblia and 13% positive for Entamoeba histolytica (cysts)
The Zoonotic and Emerging Disease group studies a range of epidemiological issues revolving around the domestic livestock, wildlife and human interface



































You must be logged in to post a comment.